Guide 5: HES demographic data (your turn!)
Script needed for this guide
- None!
Introduction
In the previous guide, we extracted a range of demographic data across children’s records on NPD. In this guide, we consider extraction of same data from HES. We might want to do this because:
- Infer missing values. You may be able to infer values from other sources, for example using NPD to complete missing records in HES. Likewise, although NPD generally has low levels of missing data on key variables, data can still be missing. Ensure you check the metadata to spot how values such as “unclear,” “not provided” or “unknown” are coded as these will not always be imported as system missing values.
- There are known differences in values across different data sources, which can have an appreciable impact on analyses. You might therefore be interested in comparing values across different datasets and running sensitivity analyses to understand how differences in coding might affect results.
- In the previous guide, we discussed how, for example, we might take modal values of time-invariant variables across a child’s records in NPD. You may wish to go one step further and compare records across NPD and HES and take modal values across these datasets.
In this guide, however, we are going to provide hints but leave it to you, as a challenge, to adapt the code we have already provided, to extract the relevant data.
The general approach
We suggest you create a new script and adopt the same general approach we took in the previous Guide. That is:
- Identify the variables you are interested in.
- Identify the HES tables you are interested in.
- Define a function that loops through these tables and extracts the relevant variables.
- Carry out necessary cleaning.
- Deduplicate and save.
Below we provide some guidance for each of these steps.
1. Identify the variables you are interested in
Let’s say you are interested in extracting data on gender/sex, ethnic group and deprivation. Your first task is to use the HES data dictionary to identify what these variables are called. The HES data dictionary is a large Excel file that can be a little clunky to navigate. There are separate sheets for each dataset (e.g., admitted patient care, outpatients and so on) and within each, each row represents one variable, sorted alphabetically. You can use Ctrl+F to search by text.
For example, Figure 1 shows the entries for IMD04 (not available in ECHILD) and IMD04_DECILE, found by searching “deprivation.”
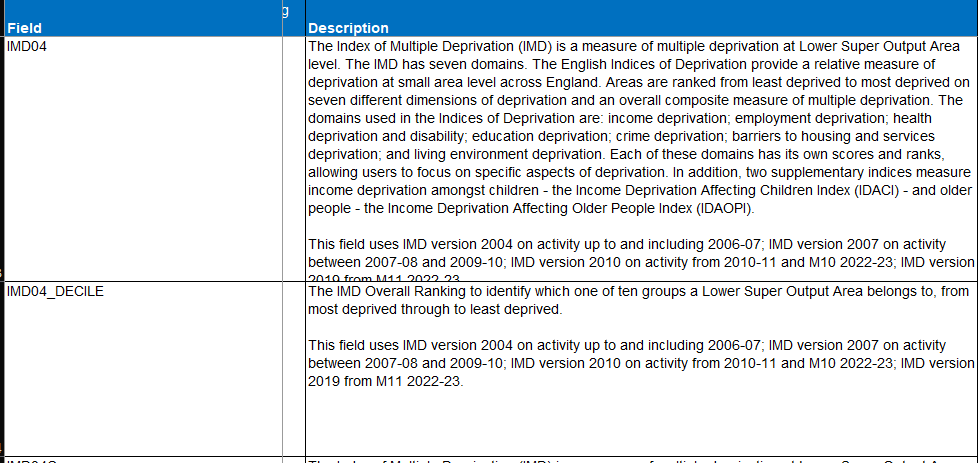
Always be careful to check not just the variable name but also its description and possible values. The IMD variable is a good example where the variable name can be misleading and researchers sometimes report this inaccurately. Although the variable is called IMD04_DECILE, suggesting it represents the 2004 edition of the IMD, the variable description reveals that different versions of the IMD are used depending on different years of activity (Figure 2).
2. Identify the HES tables you are interested in
You might want to extract data from HES admitted patient care, outpatients, emergency care or any other health dataset. In any case, for this step, you can inspect the tables available in SQL Server and/or adapt the code from Guide 3 where we extract the tables. We show an example in Code Snippet 1 in the next section. Remember, HES is organised in financial years, so ensure you select the right financial years that cover the academic years you are interested in. You can drop rows out of your date range later if you need to. You will also need to adapt the subsetting functions (i.e., changing the pattern supplied to grepl()) to ensure you select the right HES tables.
3. Define a function that loops through these tables and extracts the relevant variables
Code Snippet 1 is taken from the script that identifies our HES birth cohort (01b_identify_hes_cohort.r, Guide 3). Modifying the grepl() pattern where the tables are selected and the SQL query and supplying the correct vector of years should be enough to extract the data that you need.
Code Snippet 1 (R - 01b_identify_hes_cohort.r)
---
extract_hes_data <- function(years) {
tables <- subset(sqlTables(dbhandle), TABLE_SCHEM == "dbo")
keep <- tables$TABLE_NAME[grepl("APC", tables$TABLE_NAME)]
tables <- tables[tables$TABLE_NAME %in% keep, ]
keep <- tables$TABLE_NAME[grepl(paste0(years, collapse = "|"), tables$TABLE_NAME)]
tables <- tables[tables$TABLE_NAME %in% keep, ]
hes_source <- data.table()
for (table_name in unique(tables$TABLE_NAME)) {
gc()
print(paste0("Now doing table: ", table_name))
temp <- data.table(
sqlQuery(
dbhandle, paste0(
"SELECT TOKEN_PERSON_ID, MYDOB, EPISTART, EPIEND, ",
"ADMIDATE, DISDATE, ADMIMETH, STARTAGE, ",
"EPITYPE, CLASSPAT, NEOCARE, HRGNHS, ",
"NUMBABY, DISMETH, EPIKEY, BIRSTAT_1, MATAGE, ",
"BIRORDR_1, GESTAT_1, BIRWEIT_1, ",
paste0("DIAG_", sprintf("%02d", 1:20), collapse = ", "), ", ",
paste0("OPERTN_", sprintf("%02d", 1:24), collapse = ", "),
" FROM ", table_name)
)
)
temp <- temp[!duplicated(temp)]
hes_source <- rbind(hes_source, temp)
}
setnames(hes_source, colnames(hes_source), tolower(colnames(hes_source)))
setnames(hes_source, "token_person_id", "tokenpersonid")
return(hes_source)
}
birth_records <- extract_hes_data(vector_of_years)
---4. Carry out necessary cleaning
How you clean the variables is up to you. You at the very least need to think about missing values and ensure these are coded properly. Additional steps will then depend on how you are treating the data (i.e., as time-invariant or time-variant) and what summaries you need. Consult Guide 4 for more detail.
5. Deduplicate and save
And that’s it! See previous code for deduplication and save your dataset with an appropriate name, ready to be joined onto your spine later on.
Coming up…
The next guide, Guide 6, is only relevant if you’ve adopted an NPD inception. In it, we will look back to extract birth characteristic data. If you’ve started with a HES birth inception, you, of course, already did this when identifying your cohort and you can skip ahead to Guide 7 on health condition phenotyping.